Pipeline basics#
In case you are curious about some functionalities of MEGqc, this section provides with a short overview of some key aspects:
General Pipeline Structure#
The pipeline was initially created by Evgeniia Gaponsetva in 2023. In the following figure she described the general process of the pipeline.
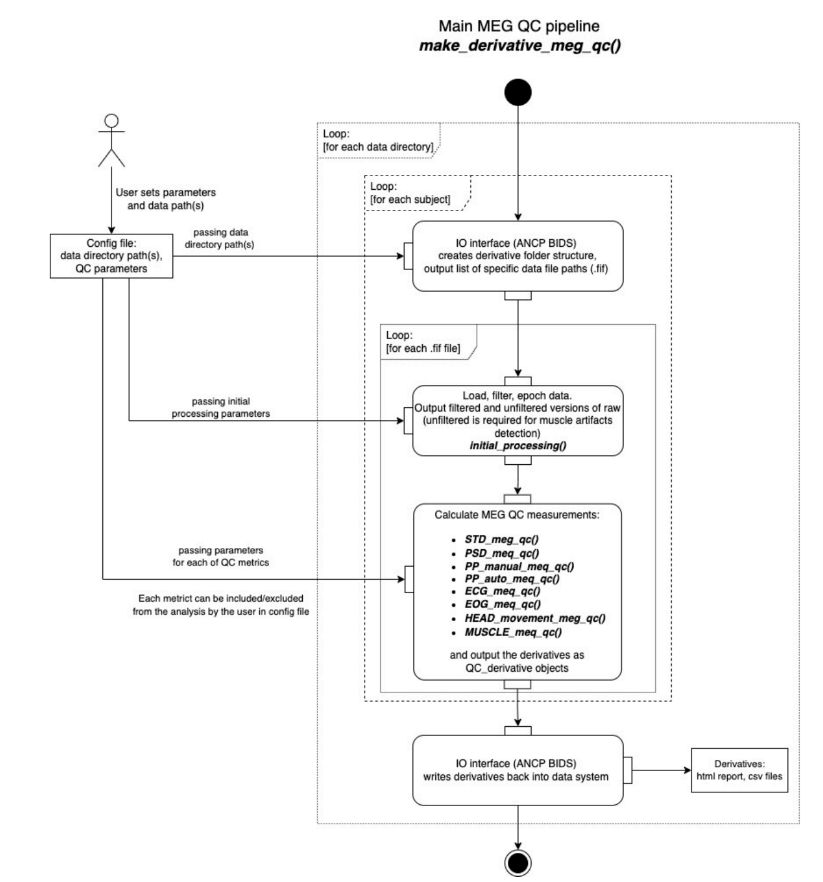
The time is on the Y axis, from top to bottom:
The configuration file contains the data directory path and the parameters for the selected metrics. Default values for all metrics are preset, but they can be modified as needed.
Data files are identified thanks to ancpBIDS.
Datasets are loaded and early processed: epoching, resampling and filtering (taking into account users’ parameters).
Selected metrics are executed (taking into account users’ parameters) and results compiled into the derivatives.
ancpBIDS writes the derivatives to the dataset directory, maintaining BIDS naming convention.
Derivatives Metadata#
BIDS suggest that the Metadata should be stored in .json and .tsv files, because both machine-readable type of file easily accesible by Python, Matlab, Excel or R. Each of the metrics of the calculation module offers both types of files:
JSON files with the key information for each of the quality metrics. JSON (JavaScript Object Notation) files are lightweight data format that store structured data in a readable, text-based format using key-value pairs, arrays, and nested objects.
TSV files with more detailed results of the metrics. The plotting module of MEGqc will use them to build the visual HTML reports. TSV (Tab-Separated Values) files are simple text files that stores tabular data, with each line representing a row and each value in the row separated by tab characters.
Dependencies#
Thanks to the installer, it’s not necessary to manually pip install the different dependencies anymore.
ancpbids==0.3.0: for BIDS compatibility and handling MEG datasets. Also developed by the ANCP Lab.mne~=1.6.0: for MEG data analysispandas>=2.0.3: data manipulation and tablesplotly=5.24.1: to create the interactive plots and visualize resultsjoblib==1.4.2: parallel processingpyqt6-tools==6.4.2.3.3: require for the graphical user interface (GUI)numba==0.58.1: compilation for performancepsutil==5.9.8: system monitoring (e.g., memory usage)matplotlib==3.8.4: plotting and figuresstatsmodels: statistical modelingscikit-learn: machine learning tools used in metricsseaborn: statistic data visualization
Want to check more extra content?
Head back to the main extra content page to explore the others!
