Calculation Module (GUI)#
Thanks to the graphical user interface (GUI) you will be able to run MEGqc and generate reports without using the terminal.
To open the GUI, click the MEGqc shortcut on your Desktop (or use the command line megqc within the environment).
(the MEGqc GUI will open alongside a bash terminal, which will display more detailed information about the on-going processes, if youc close it, the GUI will also close)
GUI visual theme
Click the left-corner icon of the GUI to open a dropdown list with all available visual themes. This will only affect the visual display of the GUI. This list also includes themes with high-contrast options.
1. Basic parameters#
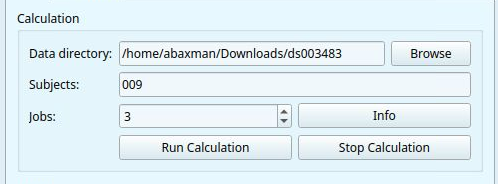
To run the calculation module you need the following basic parameters.
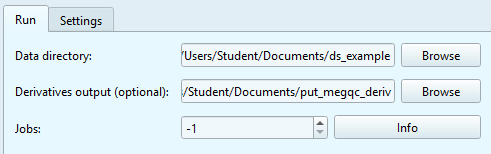
Data directory: input the root path to your dataset or click
Browseto select the folder manually. The path should lead to your whole dataset, not a particular subject.Derivatives output (optional): input the path to the folder to store the calculation output: it will create a folder with the name of the original dataset, and a “derivative” folder inside. If left blank, the output will be saved in the derivativer folder of the original dataset.
Jobs: Choose how many parallels jobs to use during the processing of your data. The default option is
1, but you can increase the speed of the processing by increasing the number of parallel Jobs.
How do I know the right amount of parallel Jobs?
MEGqc automatically detects and displays the number of cores (‘CPUs’) and the total available RAM (see the lower-corner of the GUI).
The
Infobutton will give you personalized recommendations based on system memory.You may also use
-1to use all available cores.
Before running your calculation module, you should check the Setting tab.
2. Settings#
The Setting tab allows you to customize your analysis.
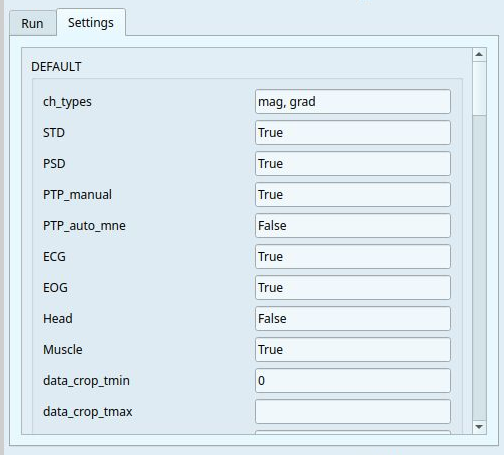
The settings are grouped into categories:
The General group of settings allows you to select the channel types (
magorgrador both), and the specific metric to compute (e.g.STD,PSDorPTP). You can also apply your analysis to a smaller snippet of data by modifyingdata_crop_tminanddata_crop_tmax.The Filtering and Epoching groups of settings allow you to edit how the filtering and epoching should be applied, such as
high-pass/low-pass cut-offs frequenciesand time windows.Metric-specfic settings: every metric includes their own editable group of settings, such as how many standard deviations from the mean to use as a threshold, the edge frequencies for PSD calculation, or the minimun PTP amplitude to count as a peak.
When you hover over each editable parameter, you’ll see a short description along with the default value. Their default values were calculated by the ANCPLab to be compatible with a broad variety of datasets.
Once you have adjust your settings, don’t forget to click on the Save Settings button at the very end of the tab.
(if you want a detailed explanation of all the settings available, visit the settings page)
3. Run Calculation#
Once you’ve set your parameters and custom your Settings, you are almost ready.
Subjects: you can enter a specific ID (e.g.,
009forsub-009). By default, all subjects will be analyzed
Finally, you can click on Run Calculation. The Log window will display the ongoing process, but you can also find more detailed information in the terminal window.